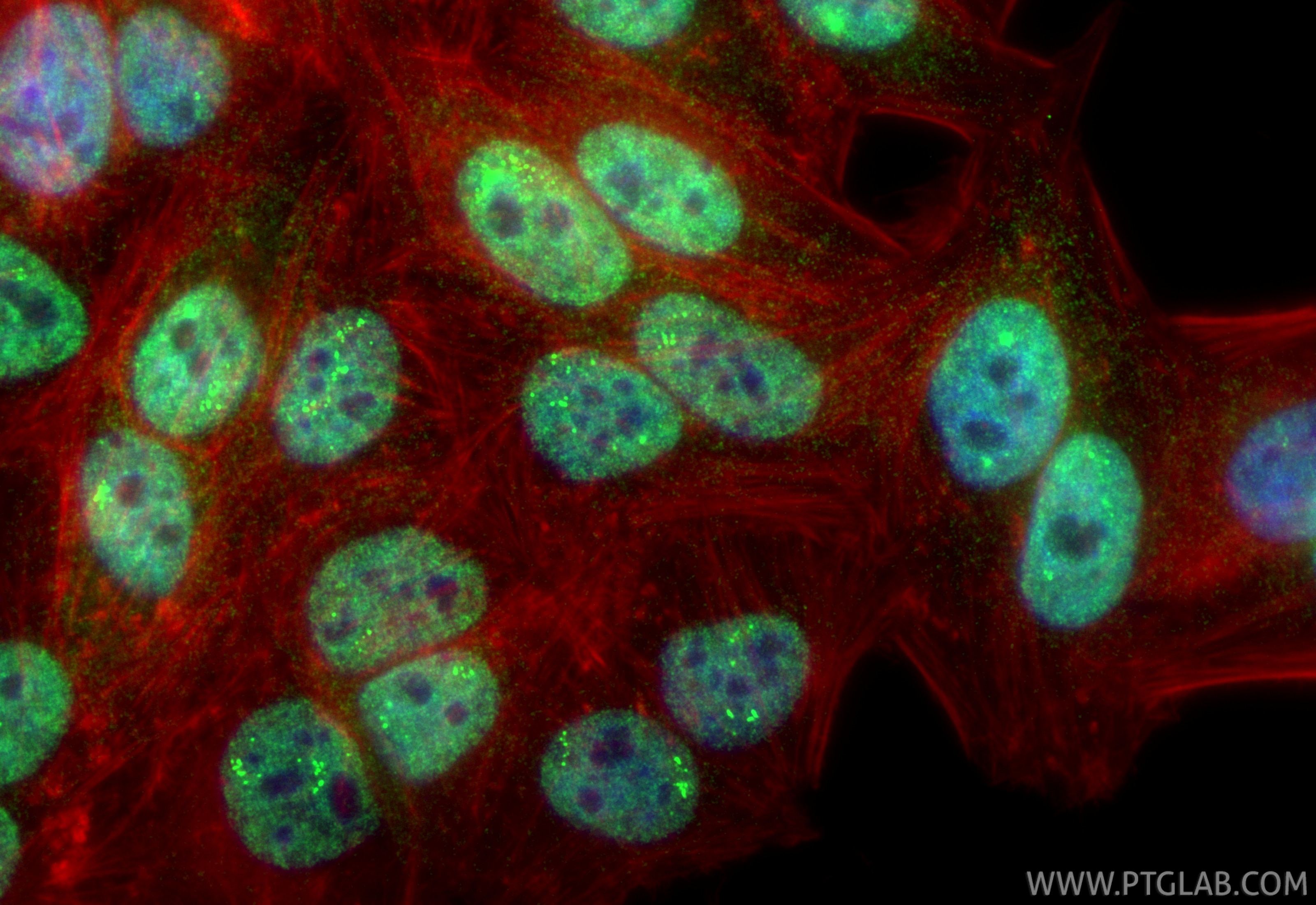
Validation Data Gallery
Tested Applications
| Positive IF/ICC detected in | HepG2 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
CL488-84823-3 targets SESN3 in IF/ICC applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen |
CatNo: Ag17872 Product name: Recombinant human SESN3 protein Source: e coli.-derived, PET28a Tag: 6*His Domain: 1-321 aa of BC017296 Sequence: MNRGGGSPSAAANYLLCTNCRKVLRKDKRIRVSQPLTRGPSAFIPEKEVVQANTVDERTNFLVEEYSTSGRLDNITQVMSLHTQYLESFLRSQFYMLRMDGPLPLPYRHYIAIMAAARHQCSYLINMHVDEFLKTGGIAEWLNGLEYVPQRLKNLNEINKLLAHRPWLITKEHIQKLVKTGENNWSLPELVHAVVLLAHYHALASFVFGSGINPERDPEISNGFRLISVNNFCVCDLANDNNIENASLSGSNFGIVDSLSELEALMERMKRLQEEREDEEASQEEMSTRFEKEKKESLFVVSGDTFHSFPHSGAFLHFFAF 相同性解析による交差性が予測される生物種 |
| Full Name | sestrin 3 |
| Calculated molecular weight | 492 aa, 57 kDa |
| Observed molecular weight | 57-60 kDa |
| GenBank accession number | BC017296 |
| Gene Symbol | SESN3 |
| Gene ID (NCBI) | 143686 |
| Conjugate | CoraLite® Plus 488 Fluorescent Dye |
| Excitation/Emission maxima wavelengths | 493 nm / 522 nm |
| Form | |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | P58005 |
| Storage Buffer | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
Sestrins, including sestrin-1 (PA26), sestrin-2 (Hi95), and sestrin-3, are 48 to 65 kDa cystein sulfinyl reductases and modulate peroxide signaling and antioxidant defense. These proteins selectively reduce or repair hyperoxidized forms of typical 2-Cys peroxiredoxins within eukaryotes. Expression of these proteins is regulated by p53, a tumor suppressor protein. Sestrin 3 was identified as a forkhead box O (FoxO) target gene with antioxidant activity. Recently it was reported that sestrin 3 may play an important role in AKT-induced increase in ROS. It might be a promising target in selectively killing cancer cells with high AKT activity levels.