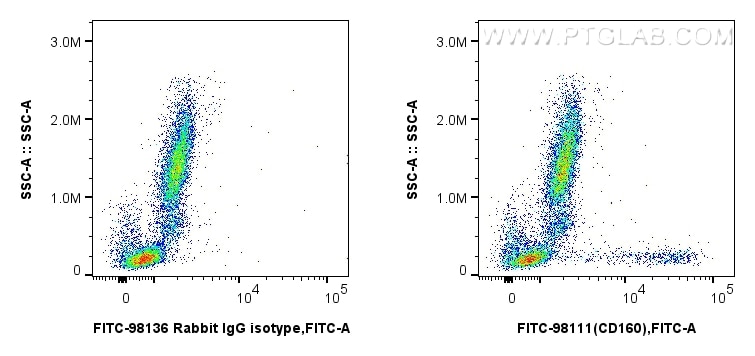
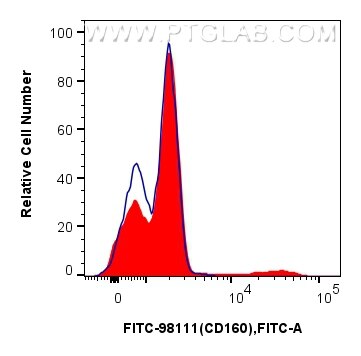
Validation Data Gallery
Tested Applications
| Positive FC detected in | human peripheral blood leukocytes |
Recommended dilution
| Application | Dilution |
|---|---|
| Flow Cytometry (FC) | FC : 5 ul per 10^6 cells in 100 μl suspension |
| This reagent has been pre-titrated and tested for flow cytometric analysis. The suggested use of this reagent is 5 ul per 10^6 cells in a 100 µl suspension or 5 ul per 100 µl of whole blood. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
FITC-98111 targets CD160 in FC applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen |
Recombinant Protein 相同性解析による交差性が予測される生物種 |
| Full Name | CD160 molecule |
| Calculated molecular weight | 20 kDa |
| GenBank accession number | BC014465 |
| Gene Symbol | CD160 |
| Gene ID (NCBI) | 11126 |
| Conjugate | FITC Plus Fluorescent Dye |
| Excitation/Emission maxima wavelengths | 495 nm / 524 nm |
| Form | |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | O95971 |
| Storage Buffer | PBS with 0.09% sodium azide and 0.5% BSA{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at 2-8°C. Avoid exposure to light. Stable for one year after shipment. |
Background Information
CD160 is a glycosylphosphatidylinositol (GPI)-anchored protein that is expressed on almost all intestinal intraepithelial lymphocytes (IELs), gamma delta T cells, CD56 (dim) CD16+ cytotoxic NK cells, and a minor subset of CD4+ and CD8+ T cells (PMID: 31332204; 25179432). The binding of CD160 to both classical and non-classical MHC I enhances NK and CD8+ cytotoxic T lymphocyte functions (PMID: 25179432). CD160 is a ligand for HVEM (herpes virus entry mediator) and the binding of CD160 to HVEM inhibits T cell activity (PMID: 19426226).
Protocols
| Product Specific Protocols | |
|---|---|
| FC protocol for FITC Plus CD160 antibody FITC-98111 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |