Validation Data Gallery
Tested Applications
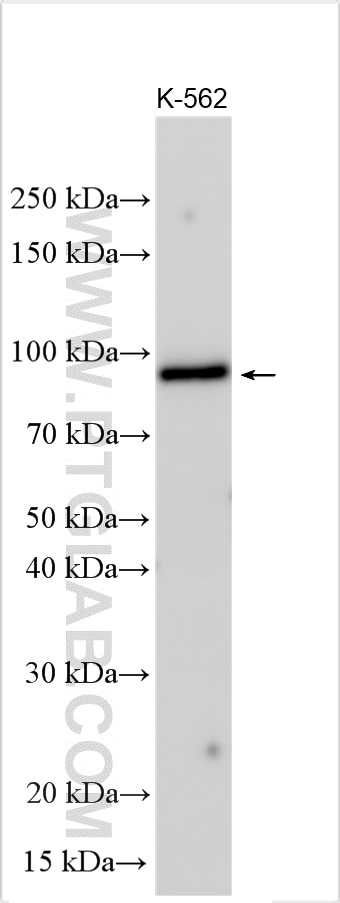
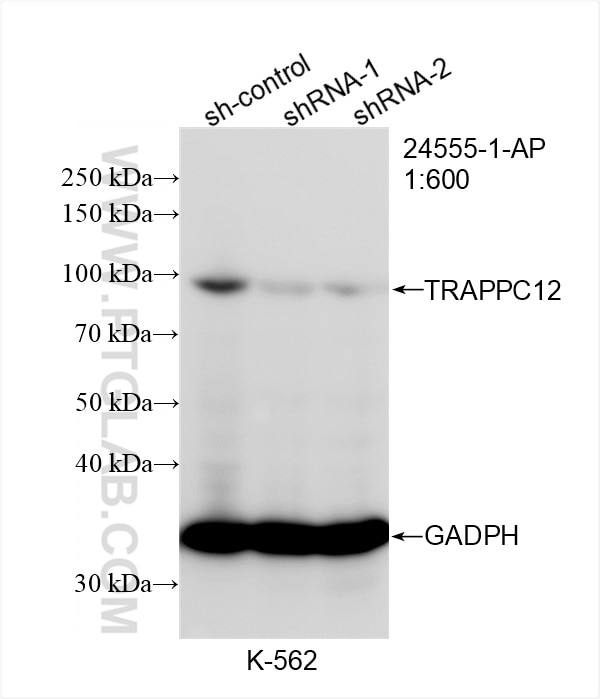
| Positive WB detected in | K-562 cells |
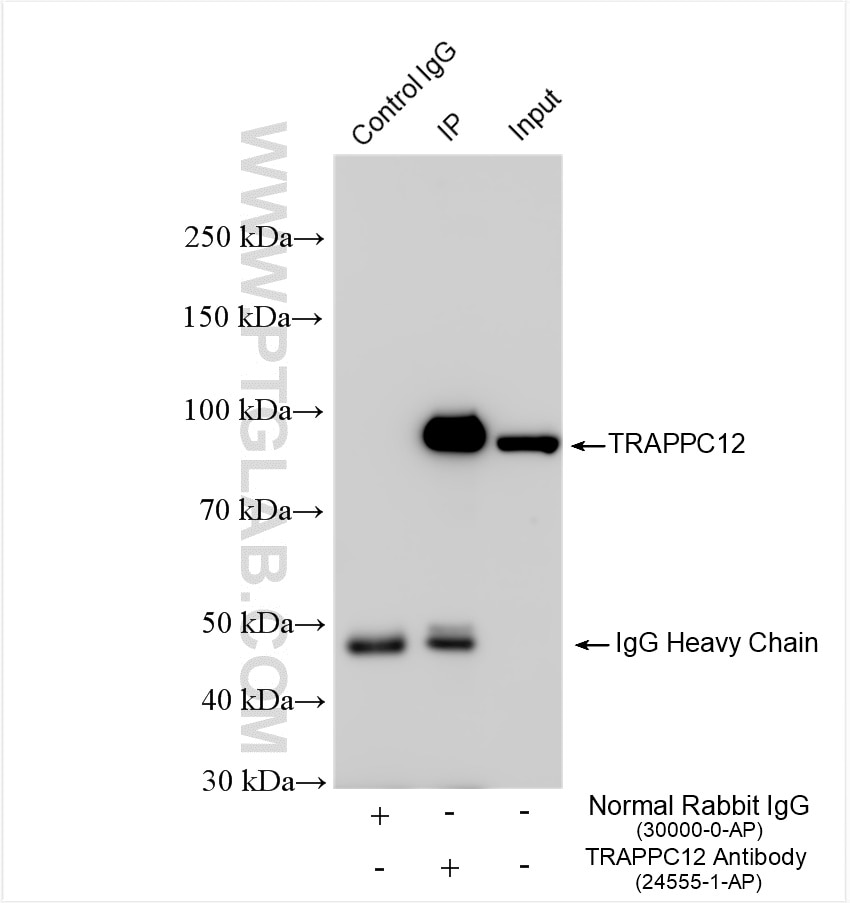
| Positive IP detected in | K-562 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:500-1:2000 |
| Immunoprecipitation (IP) | IP : 0.5-4.0 ug for 1.0-3.0 mg of total protein lysate |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
24555-1-AP targets TRAPPC12 in WB, IP, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen | TRAPPC12 fusion protein Ag18332 相同性解析による交差性が予測される生物種 |
| Full Name | tetratricopeptide repeat domain 15 |
| Calculated molecular weight | 735 aa, 79 kDa |
| Observed molecular weight | 98 kDa |
| GenBank accession number | BC014164 |
| Gene Symbol | TRAPPC12 |
| Gene ID (NCBI) | 51112 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q8WVT3 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
TRAPPC12, also called CGI-87, TTC15 and TRAMM, has been recently identified as subunit of TRAPP complex in two independent proteomic studies. It was suggested that TRAPPC12 was a subunit of TRAPPIII. TRAPPC12 and TECPR1 acted at distinct steps in autophagy and depletion of TRAPPC12 and/or TECPR1 increased in the number of autophagosomes and increase autophagic flux. The observed molecular weight of TRAPPC12 is 98 kDa, which is consistent with what has been described in the literature (PMID: 28240221, PMID: 38439956).
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for TRAPPC12 antibody 24555-1-AP | Download protocol |
| IP protocol for TRAPPC12 antibody 24555-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |