Validation Data Gallery
Tested Applications
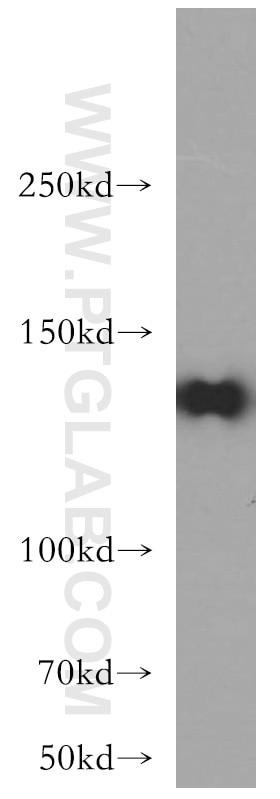
| Positive WB detected in | L02 cells |
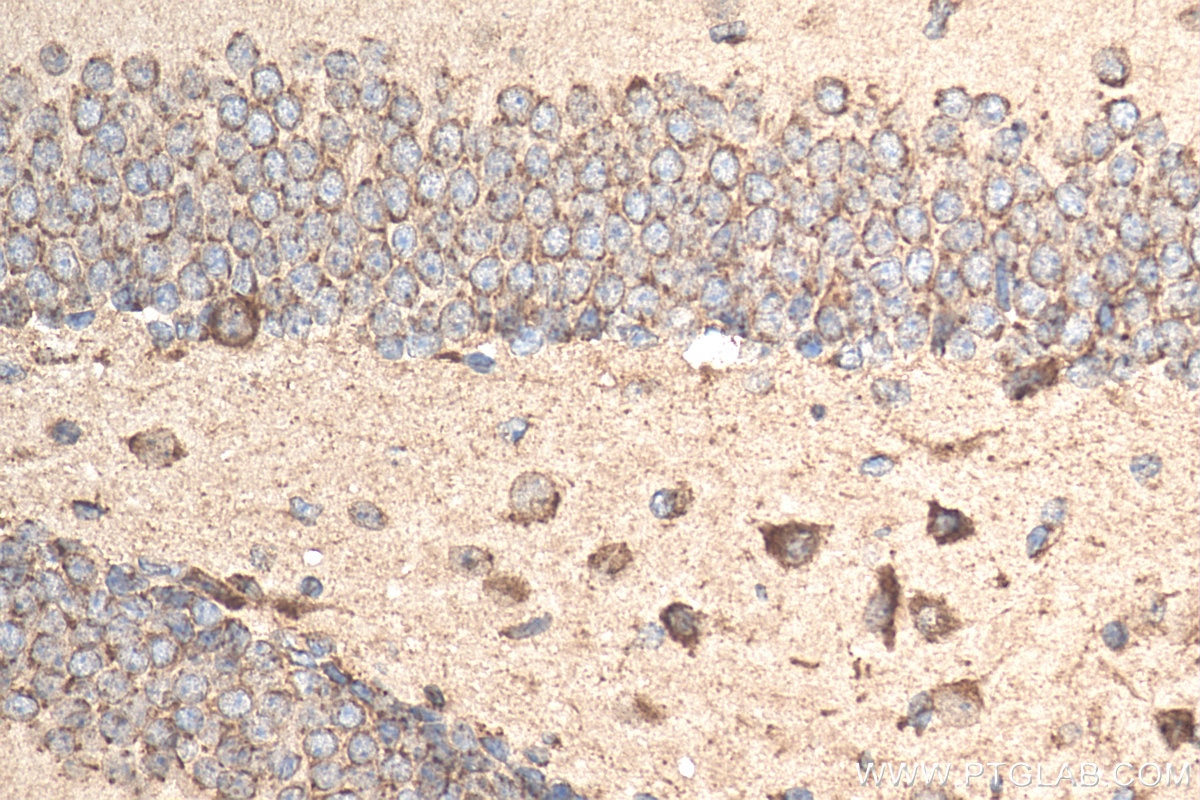
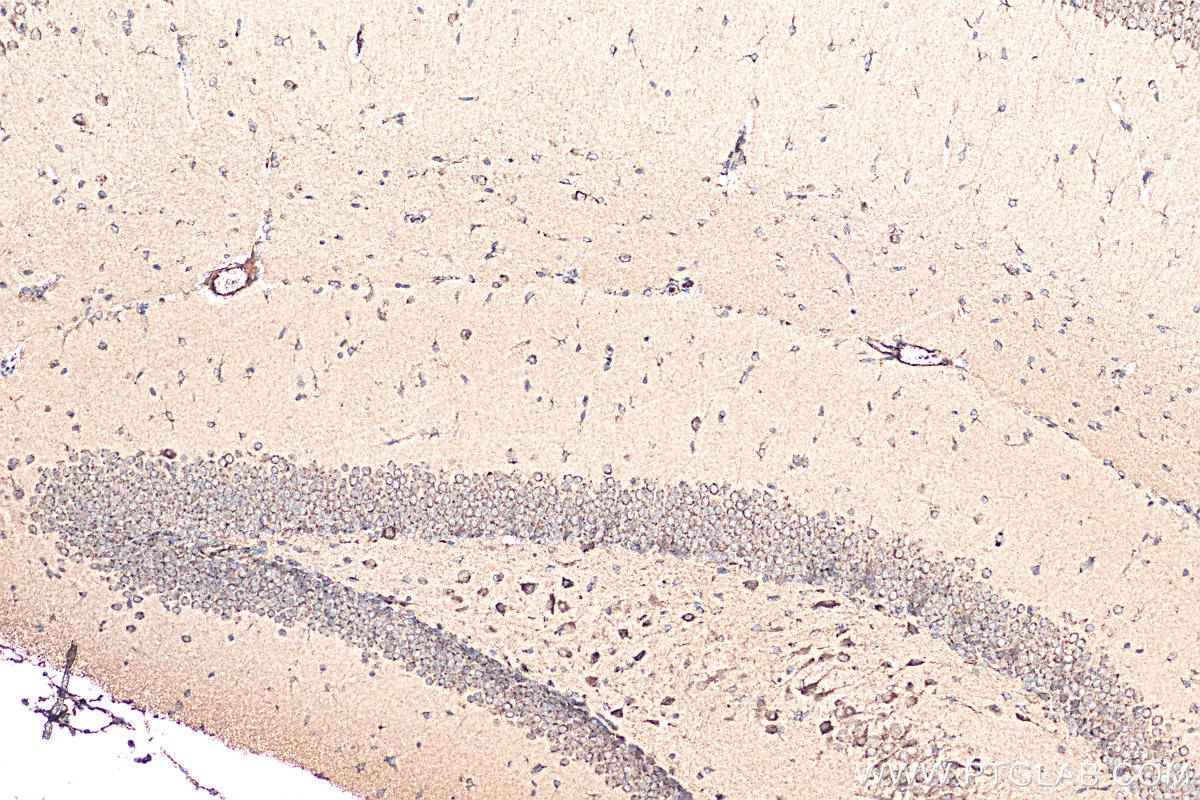
| Positive IHC detected in | mouse brain tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:500-1:2000 |
| Immunohistochemistry (IHC) | IHC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Published Applications
| WB | See 1 publications below |
Product Information
20093-1-AP targets RIM2-Specific in WB, IHC, ELISA applications and shows reactivity with human, mouse samples.
| Tested Reactivity | human, mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
Peptide 相同性解析による交差性が予測される生物種 |
| Full Name | regulating synaptic membrane exocytosis 2 |
| Calculated molecular weight | 160 kDa |
| Observed molecular weight | 133 kDa |
| GenBank accession number | NM_001100117 |
| Gene Symbol | RIMS2 |
| Gene ID (NCBI) | 9699 |
| RRID | AB_10666429 |
| Conjugate | Unconjugated |
| Form | |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q9UQ26 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
RIMS2, also named as KIAA0751, RAB3IP3 and RIM2, is a rab effector which involved in exocytosis. RIMS2 may act as scaffold protein. This antibody is specific to RIMS2. It recognizes all the isoforms except isoform7 of RIMS2.
Protocols
| Product Specific Protocols | |
|---|---|
| IHC protocol for RIM2-Specific antibody 20093-1-AP | Download protocol |
| WB protocol for RIM2-Specific antibody 20093-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |