Validation Data Gallery
Tested Applications
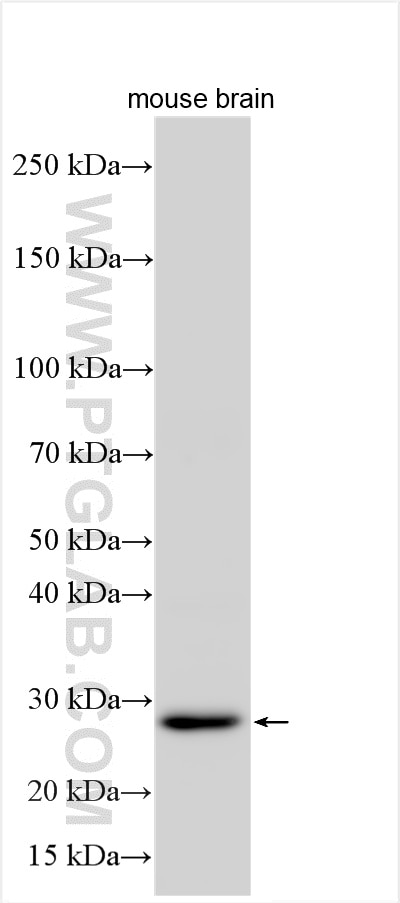
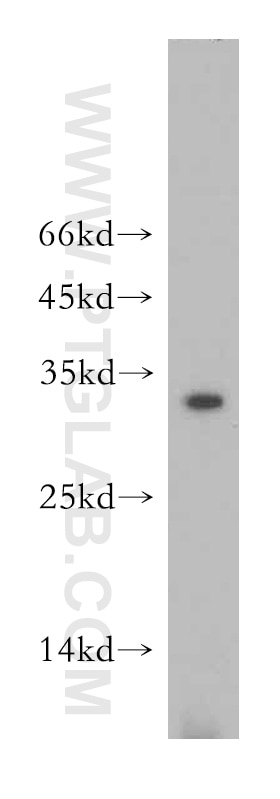
| Positive WB detected in | mouse brain tissue, mouse skeletal muscle tissue |
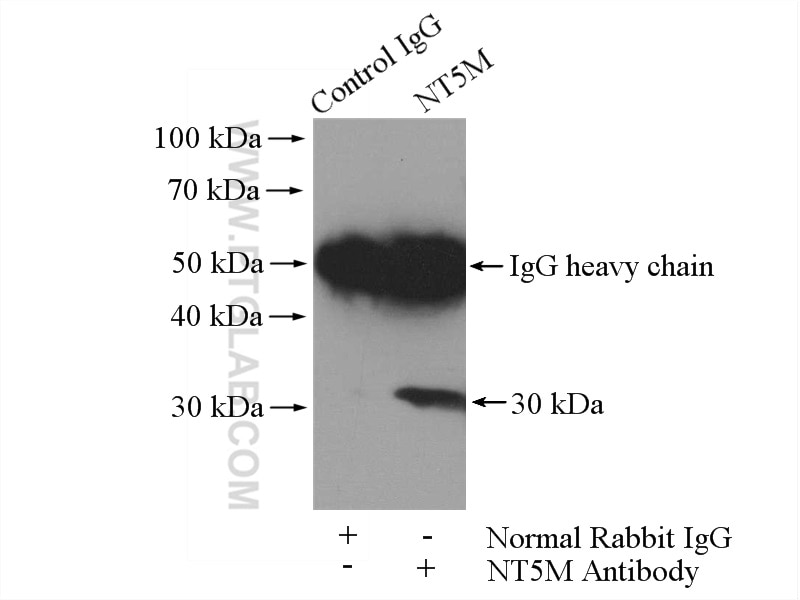
| Positive IP detected in | mouse skeletal muscle tissue |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:1000-1:6000 |
| Immunoprecipitation (IP) | IP : 0.5-4.0 ug for 1.0-3.0 mg of total protein lysate |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
20765-1-AP targets NT5M in WB, IP, ELISA applications and shows reactivity with mouse samples.
| Tested Reactivity | mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag14743 Product name: Recombinant human NT5M protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 36-228 aa of BC035838 Sequence: LRVLVDMDGVLADFEGGFLRKFRARFPDQPFIALEDRRGFWVSEQYGRLRPGLSEKAISIWESKNFFFELEPLPGAVEAVKEMASLQNTDVFICTSPIKMFKYCPYEKYAWVEKYFGPDFLEQIVLTRDKTVVSADLLIDDRPDITGAEPTPSWEHVLFTACHNQHLQLQPPRRRLHSWADDWKAILDSKRPC 相同性解析による交差性が予測される生物種 |
| Full Name | 5',3'-nucleotidase, mitochondrial |
| Calculated molecular weight | 228 aa, 26 kDa |
| Observed molecular weight | 26-30 kDa |
| GenBank accession number | BC035838 |
| Gene Symbol | NT5M |
| Gene ID (NCBI) | 56953 |
| RRID | AB_10733338 |
| Conjugate | Unconjugated |
| Form | |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q9NPB1 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
NT5M is a mitochondrial 5′(3′)-deoxyribonucleotidase that plays a crucial role in maintaining balanced deoxynucleotide pools essential for mitochondrial DNA synthesis. It dephosphorylates specific nucleoside monophosphates, particularly dUMP and dTMP, thereby preventing the accumulation of toxic dTTP. NT5M is predominantly localized in mitochondria and is highly expressed in tissues such as the heart, brain, and skeletal muscle.
Protocols
| Product Specific Protocols | |
|---|---|
| IP protocol for NT5M antibody 20765-1-AP | Download protocol |
| WB protocol for NT5M antibody 20765-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |