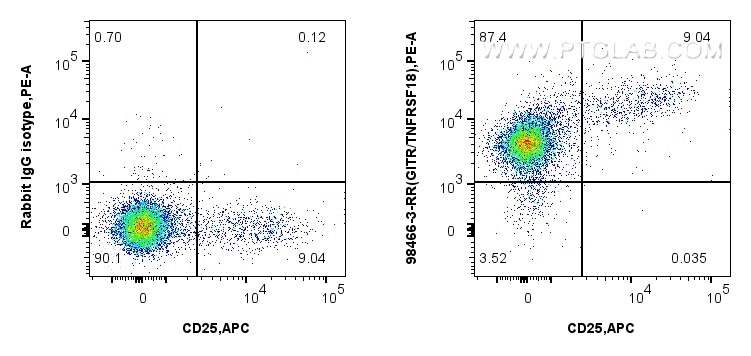
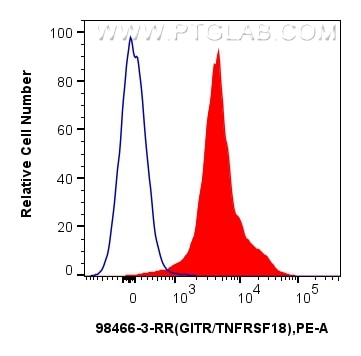
Validation Data Gallery
Tested Applications
| Positive FC detected in | mouse splenocytes |
Recommended dilution
| Application | Dilution |
|---|---|
| Flow Cytometry (FC) | FC : 0.25 ug per 10^6 cells in a 100 µl suspension |
| This reagent has been tested for flow cytometric analysis. It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
98466-3-RR targets GITR/TNFRSF18 in FC applications and shows reactivity with mouse samples.
| Tested Reactivity | mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | GITR/TNFRSF18 fusion protein Eg3695 相同性解析による交差性が予測される生物種 |
| Full Name | tumor necrosis factor receptor superfamily, member 18 |
| Calculated molecular weight | 25kDa |
| GenBank accession number | NM_009400.2 |
| Gene Symbol | CD357 |
| Gene ID (NCBI) | 21936 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | O35714-1 |
| Storage Buffer | PBS with 0.09% sodium azide{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at 2 - 8°C. Stable for one year after shipment. |
Background Information
Glucocorticoid-induced TNFR-related protein (GITR), also known as CD357 or TNFRSF18, is a member of the tumor necrosis factor receptor (TNF-R) superfamily. GITR is expressed constitutively at high levels in T regulatory cells (Treg cells) and plays a key role in dominant immunological self-tolerance maintained by CD25+CD4+ regulatory T cells (PMID: 11812990). It is expressed at low levels on resting responder T cells. The expression of GITR on T cells can be upregulated upon activation (PMID: 15770698). GITR is activated by GITR ligand (GITRL) which is mainly expressed on APC. GITR-GITRL interactions could co-stimulate both responder T-cell functions and the suppressive functions of Treg cells (PMID: 16868552).
Protocols
| Product Specific Protocols | |
|---|---|
| FC protocol for GITR/TNFRSF18 antibody 98466-3-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |