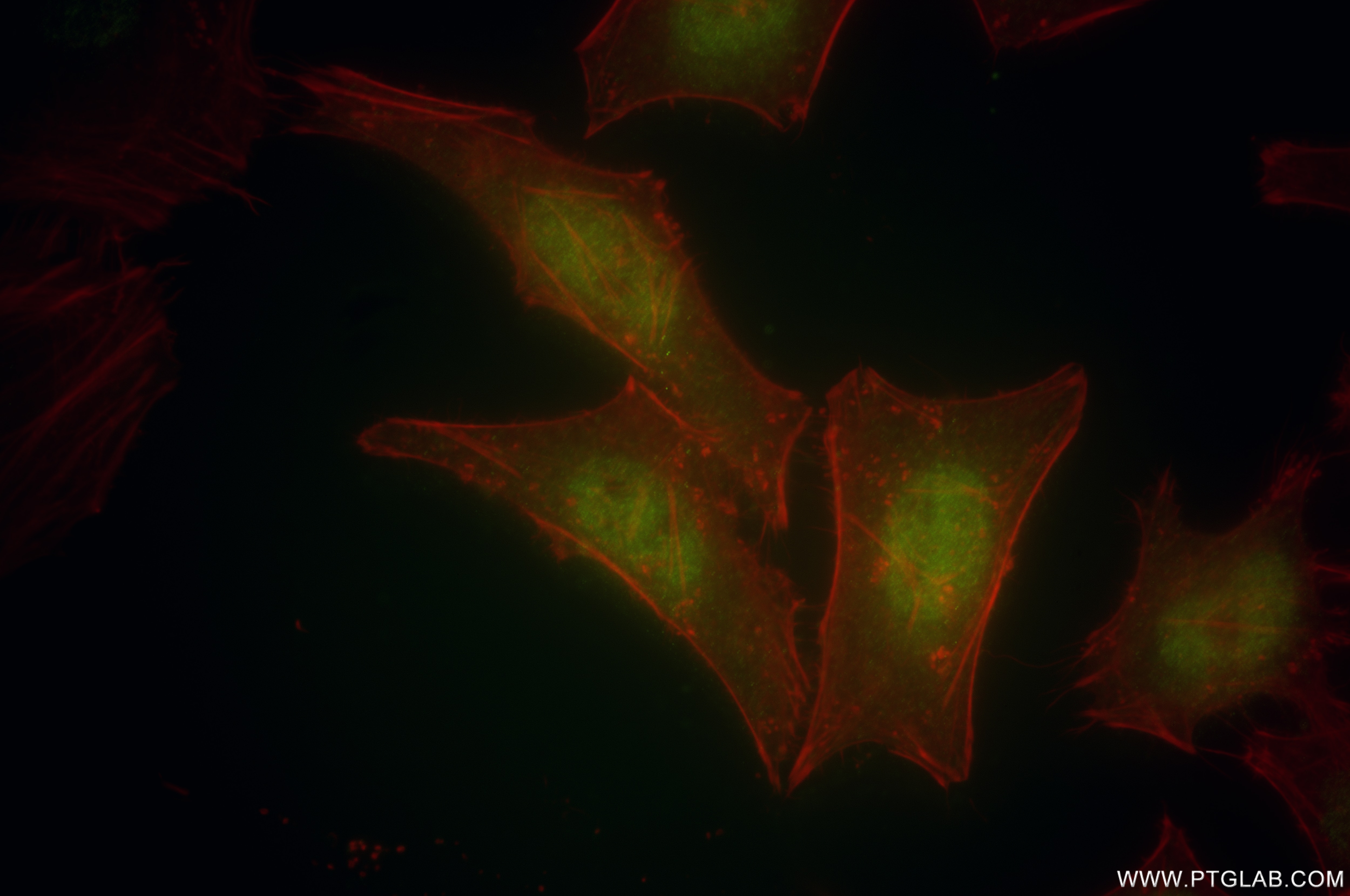
Validation Data Gallery
Tested Applications
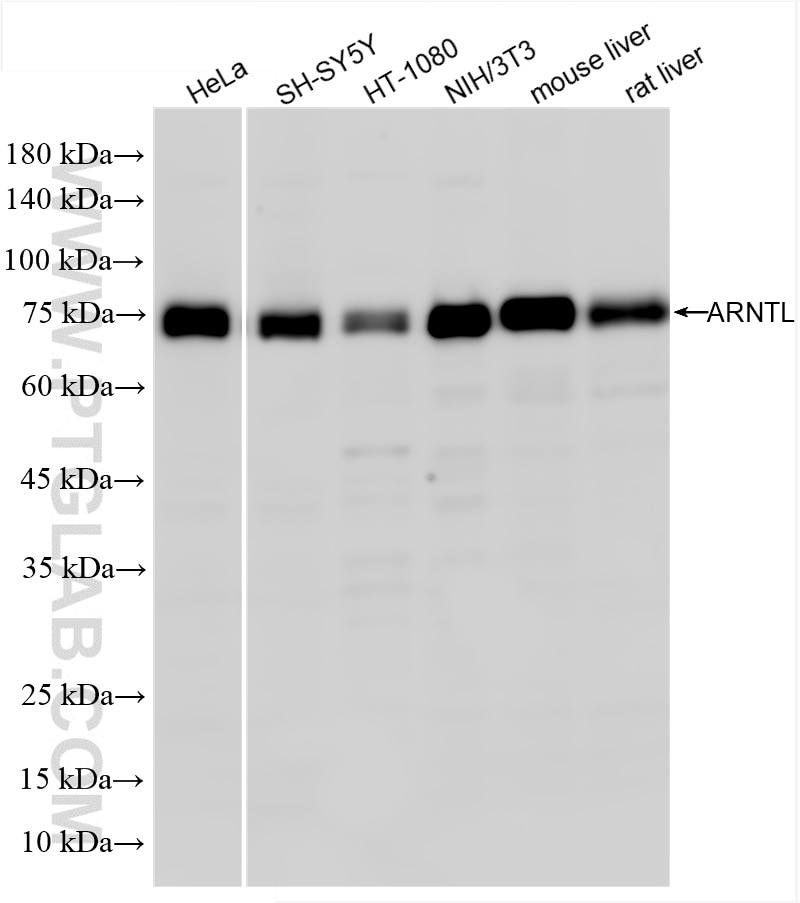
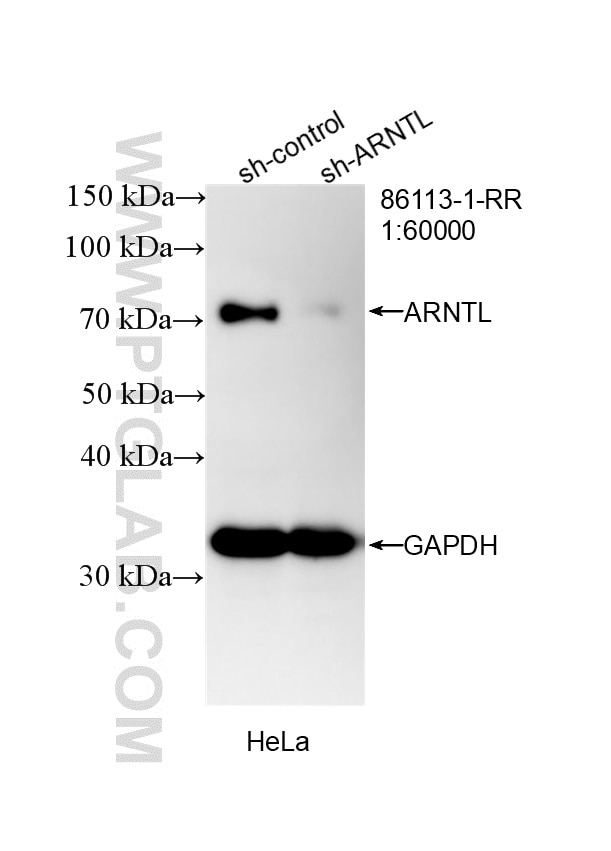
| Positive WB detected in | HeLa cells, SH-SY5Y cells, HT-1080 cells, NIH/3T3 cells, mouse liver tissue, rat liver tissue |
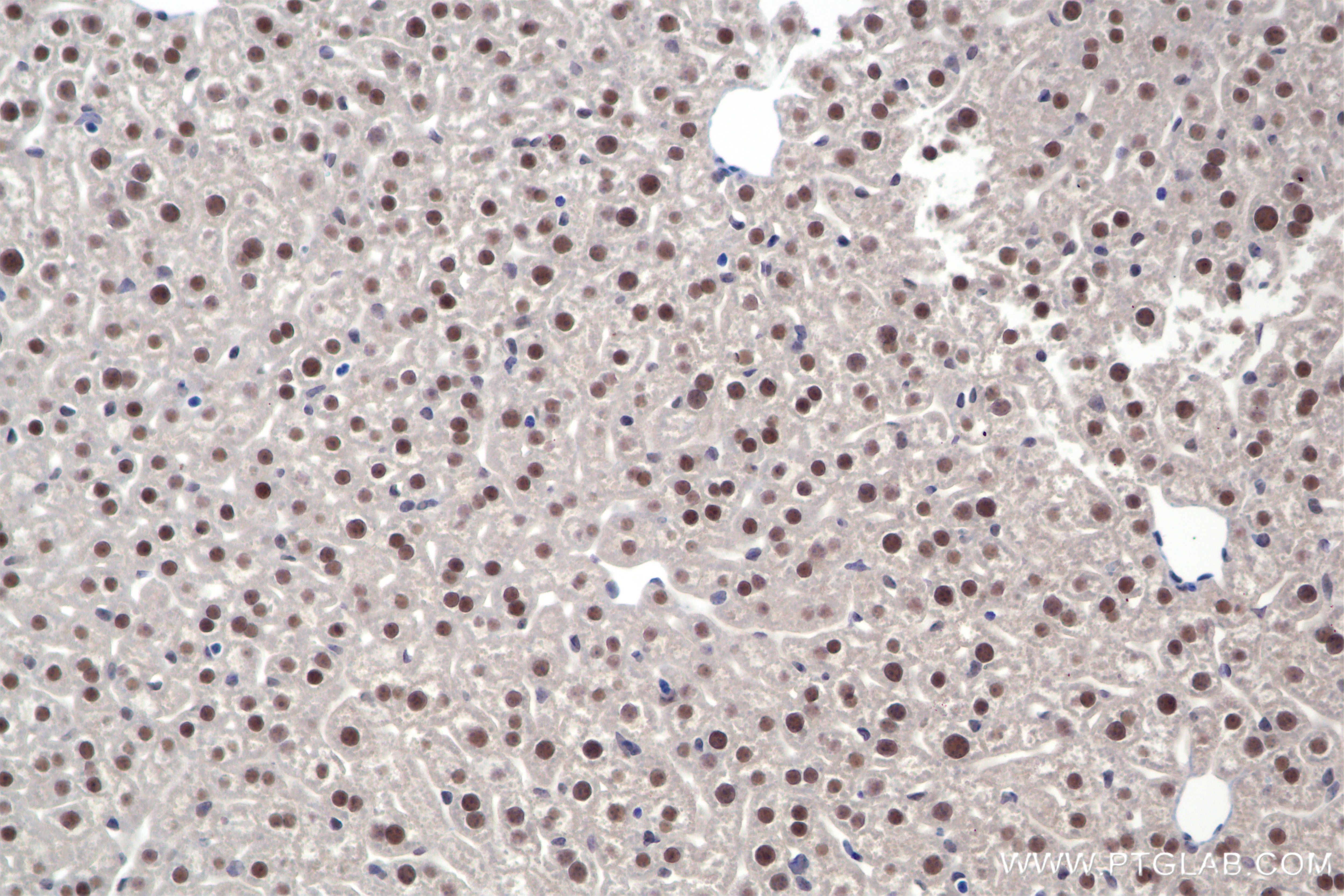
| Positive IHC detected in | mouse liver tissue, rat liver tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
| Positive IF/ICC detected in | HeLa cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Immunohistochemistry (IHC) | IHC : 1:1000-1:4000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:250-1:1000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
86113-1-RR targets ARNTL in WB, IHC, IF/ICC, ELISA applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen |
CatNo: Ag5586 Product name: Recombinant human ARNTL protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-303 aa of BC041129 Sequence: MADQRMDISSTISDFMSPGPTDLLSSSLGTSGVDCNRKRKGSSTDYQESMDTDKDDPHGRLEYTEHQGRIKNAREAHSQIEKRRRDKMNSFIDELASLVPTCNAMSRKLDKLTVLRMAVQHMKTLRGATNPYTEANYKPTFLSDDELKHLILRAADGFLFVVGCDRGKILFVSESVFKILNYSQNDLIGQSLFDYLHPKDIAKVKEQLSSSDTAPRERLIDAKTGLPVKTDITPGPSRLCSGARRSFFCRMKCNRPSVKVEDKDFPSTCSKKKDRKSFCTIHSTGYLKSWPPTKMGLDEDNEP 相同性解析による交差性が予測される生物種 |
| Full Name | aryl hydrocarbon receptor nuclear translocator-like |
| Calculated molecular weight | 69 kDa |
| Observed molecular weight | 69-75 kDa |
| GenBank accession number | BC041129 |
| Gene Symbol | ARNTL |
| Gene ID (NCBI) | 406 |
| Conjugate | Unconjugated |
| Form | |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | O00327 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
BMAL1, a basic helix-loop-helices with Per-Arnt-Sim (PAS) domain transcription factor, is one of components that has core clock function in mammals. It form a heterodimer with CLOCK, and induce the transcription of target genes by binding to E-box elements within their promoter regions. And this transcription is inhibited in a feedback loop by PER, and also by CRY proteins.Also BMAL1 is a key player in the regulation of metabolism, playing a role in adipogenesis and in the control of glucose and triglycerides levels. Due to phosphorylation modification, BMAL1 can be detected as a band in the range of 70-80 kda (PMID: 14750952).
Protocols
| Product Specific Protocols | |
|---|---|
| IHC protocol for ARNTL antibody 86113-1-RR | Download protocol |
| WB protocol for ARNTL antibody 86113-1-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |