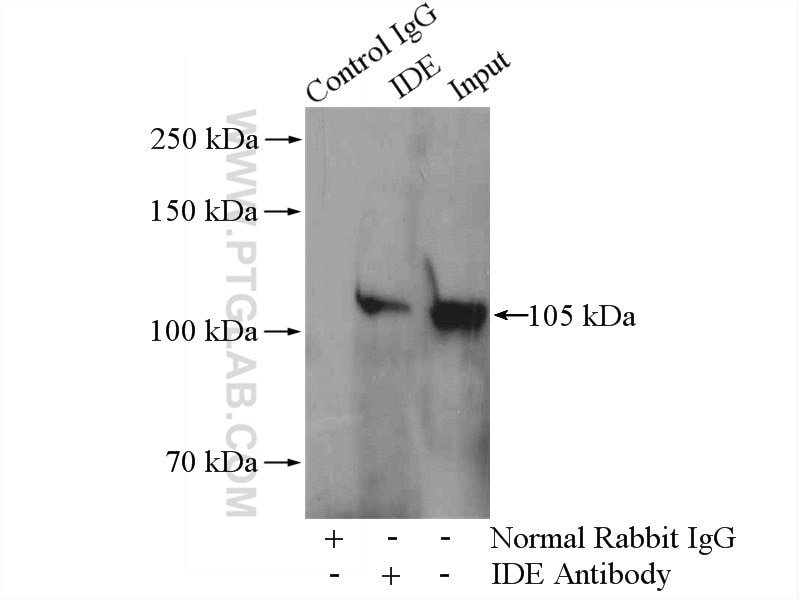
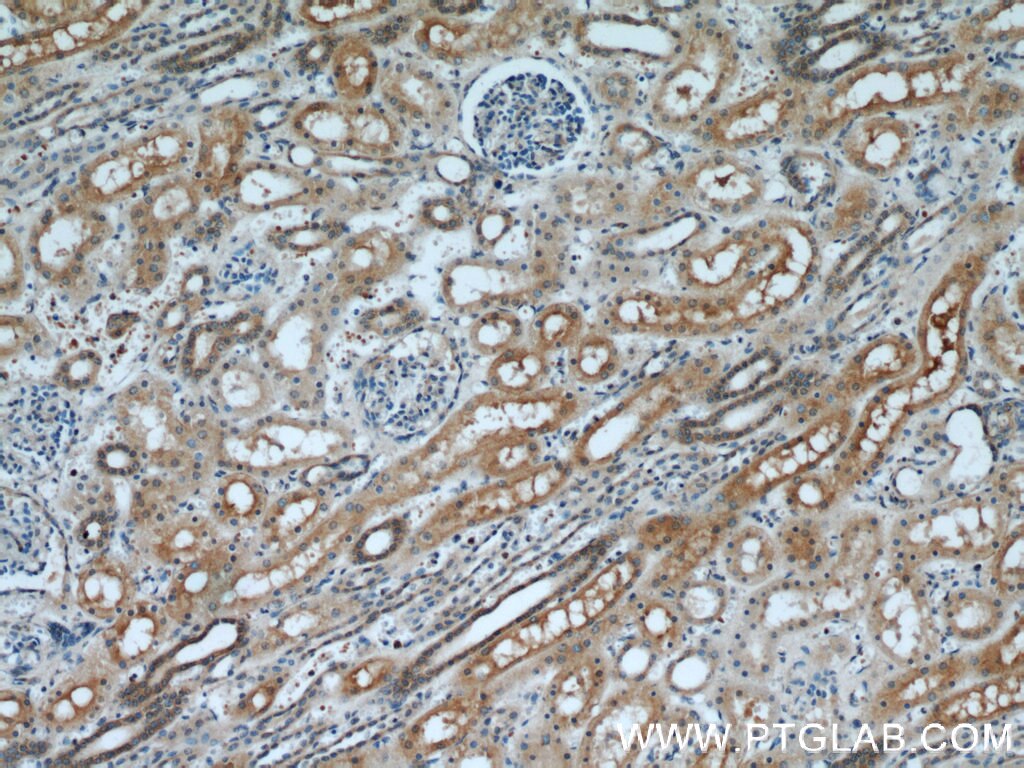
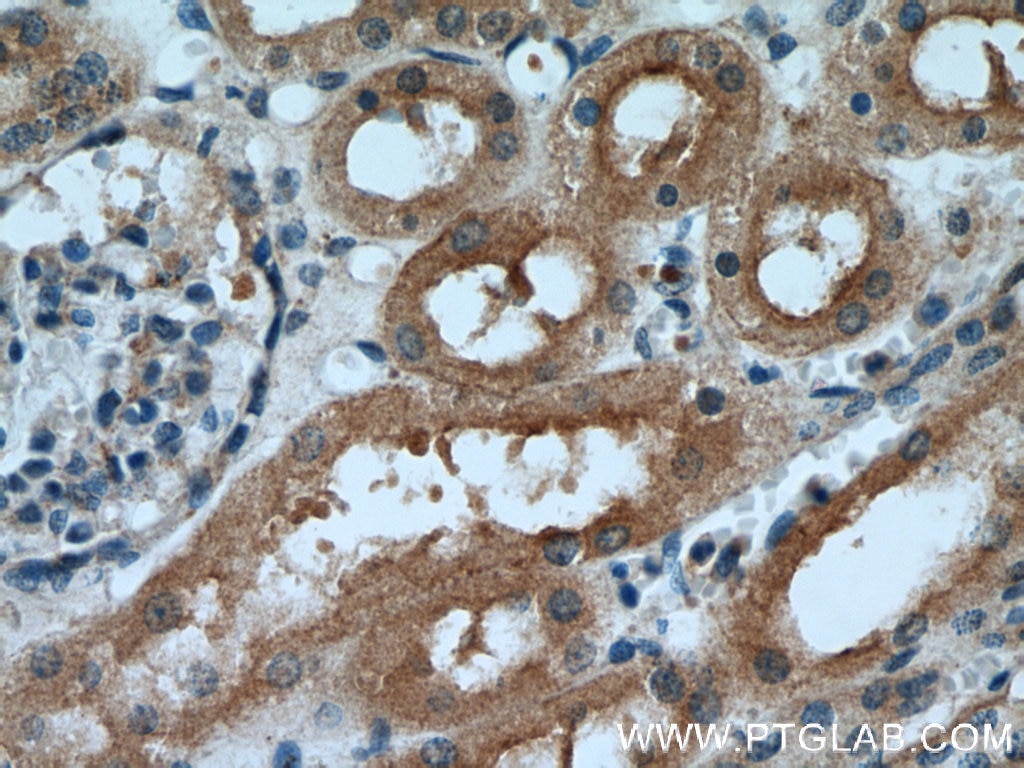
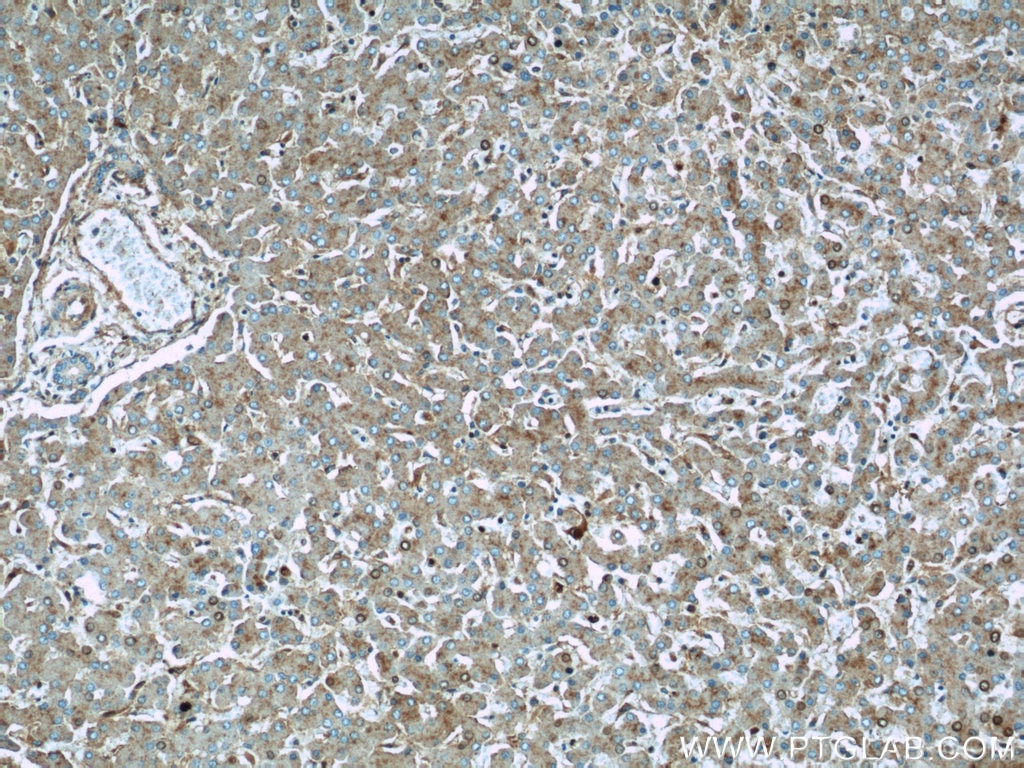
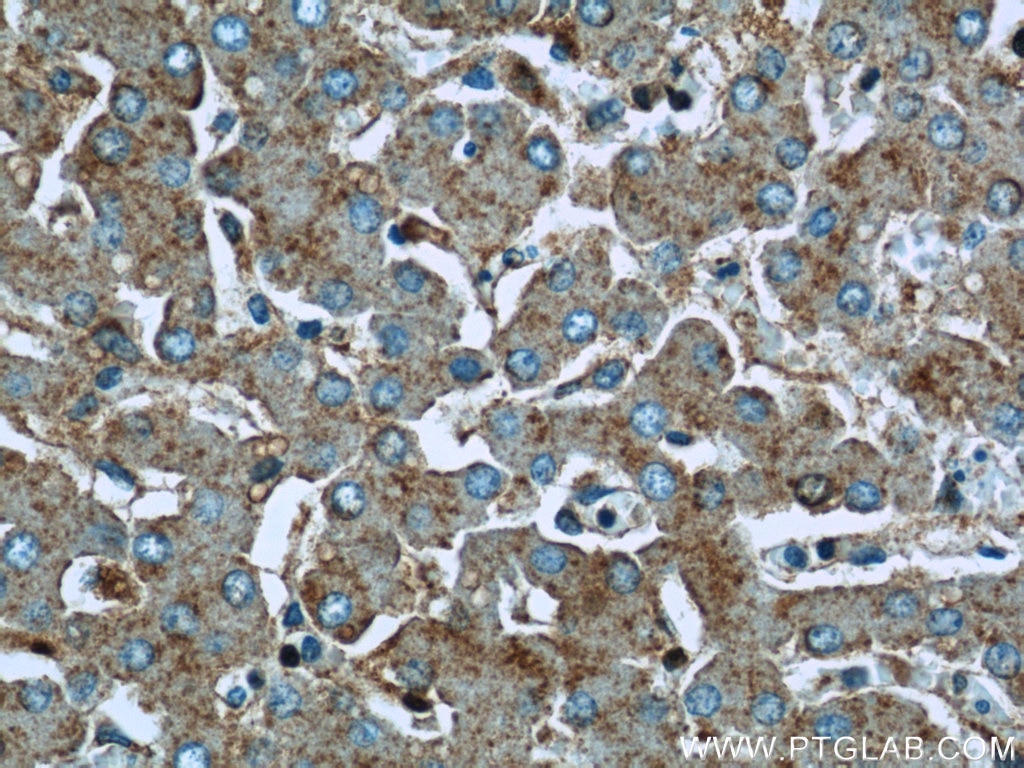
Validation Data Gallery
Tested Applications
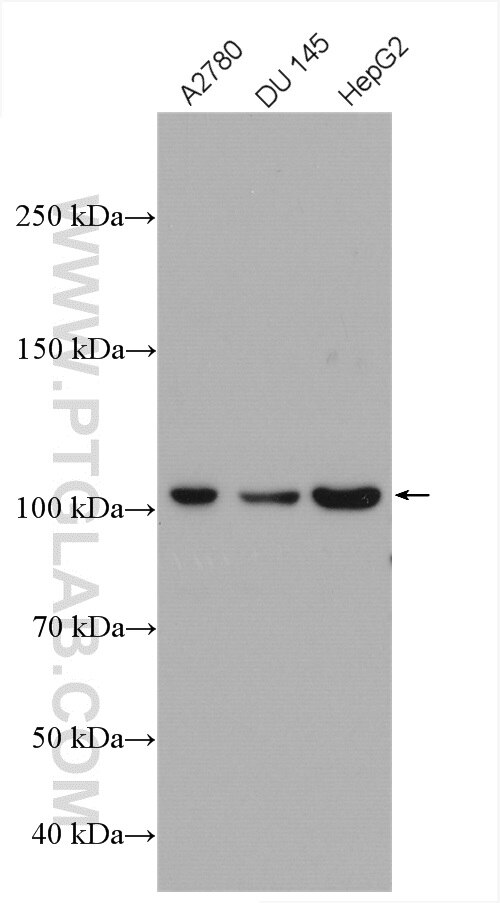
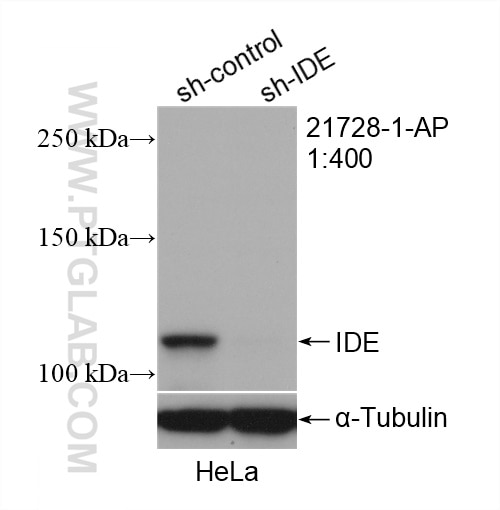
| Positive WB detected in | A2780 cells, HepG2 cells, HeLa cells, DU 145 cells |
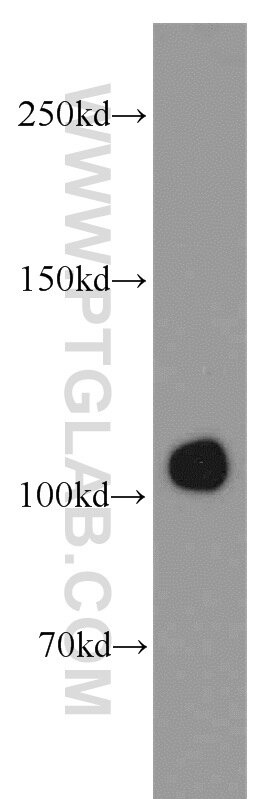
| Positive IP detected in | HepG2 cells |
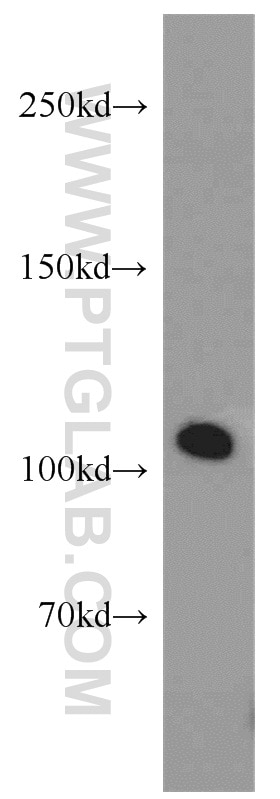
| Positive IHC detected in | human kidney tissue, human liver tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:500-1:1000 |
| Immunoprecipitation (IP) | IP : 0.5-4.0 ug for 1.0-3.0 mg of total protein lysate |
| Immunohistochemistry (IHC) | IHC : 1:20-1:200 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Published Applications
| WB | See 12 publications below |
| IHC | See 1 publications below |
Product Information
21728-1-AP targets IDE in WB, IHC, IP, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Cited Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag16392 Product name: Recombinant human IDE protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 676-1019 aa of BC096336 Sequence: EQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIPQLLSRLHIEALLHGNITKQAALGIMQMVEDTLIEHAHTKPLLPSQLVRYREVQLPDRGWFVYQQRNEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQGLRFIIQSEKPPHYLESRVEAFLITMEKSIEDMTEEAFQKHIQALAIRRLDKPKKLSAECAKYWGEIISQQYNFDRDNTEVAYLKTLTKEDIIKFYKEMLAVDAPRRHKVSVHVLAREMDSCPVVGEFPCQNDINLSQAPALPQPEVIQNMTEFKRGLPLFPLVKPHINFMAAKL 相同性解析による交差性が予測される生物種 |
| Full Name | Abeta degrading protease |
| Calculated molecular weight | 1019 aa, 118 kDa |
| Observed molecular weight | 100-120 kDa |
| GenBank accession number | BC096336 |
| Gene Symbol | IDE |
| Gene ID (NCBI) | 3416 |
| RRID | AB_10804773 |
| Conjugate | Unconjugated |
| Form | |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | P14735 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
IDE (INS-degrading enzyme) belongs to the peptidase M16 family. It is also known as insulysin and is a 110-kD neutral metallopeptidase that can degrade a number of peptides, including INS and beta-amyloid(PMID:9830016). The reduction of IDE expression may be a risk factor for AD, and that IDE may interact with APOE status to affect beta-amyloid metabolism(PMID:12507914). It can exsit as a homodimer and form higher oligomers(PMID:17613531). This antibody is specific to IDE.
Protocols
| Product Specific Protocols | |
|---|---|
| IHC protocol for IDE antibody 21728-1-AP | Download protocol |
| IP protocol for IDE antibody 21728-1-AP | Download protocol |
| WB protocol for IDE antibody 21728-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |
Publications
| Species | Application | Title |
|---|---|---|
Brain MMP13 inhibition rescues cognitive decline in Alzheimer transgenic mice via BACE1 regulation. | ||
Carbohydr Polym A pectin from fruits of Lycium barbarum L. decreases β-amyloid peptide production through modulating APP processing. | ||
Cell Physiol Biochem Novel Serum Biomarkers Detected by Protein Array in Polycystic Ovary Syndrome with Low Progesterone Level. | ||
Int J Biol Macromol A novel pectin from Polygala tenuifolia blocks Aβ42 aggregation and production by enhancing insulin-degradation enzyme and neprilysin. | ||
J Biol Chem Cell surface expression of the major amyloid-β peptide (Aβ)-degrading enzyme, neprilysin, depends on phosphorylation by mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK) and dephosphorylation by protein phosphatase 1a. | ||
Neurochem Int Activation of α7 nicotinic acetylcholine receptor alleviates Aβ1-42-induced neurotoxicity via downregulation of p38 and JNK MAPK signaling pathways. |