RIP/RIP-seq to study RNA Modifications
Written by Sripriya Raja, PhD Candidate at University of Pittsburgh.
What is RIP/RIP-seq?
RIP/RIP-seq, or RNA immunoprecipitation/RNA immunoprecipitation sequencing, is a high-throughput experimental technique used to study the interaction between RNA and RNA-binding proteins. It allows for the identification of specific RNA-bound proteins within cells or tissues.
Why is it beneficial to study RNA modifications?
Studying RNA modifications is essential for unraveling the complexity of RNA biology, understanding gene expression regulation, deciphering disease mechanisms, and exploring diagnostic and therapeutic applications. It provides a deeper level of understanding of RNA molecules and their roles in cellular processes and diseases.
The dysregulation of RNA modifications has been implicated in various diseases, including cancer, neurological disorders, and metabolic disorders. Understanding RNA modifications opens opportunities for developing targeted therapeutic approaches. By manipulating the enzymes responsible for adding or erasing specific RNA modifications, it may be possible to modulate gene expression, alter RNA structure, or correct aberrant RNA modifications in disease states. Additionally, modulating RNA modifications pharmacologically may offer new avenues for therapeutic intervention. Understanding the landscape of RNA modifications and their functional consequences can guide the development of RNA-targeted therapies and precision medicine approaches.
RIP/RIP-seq provides key insights into the binding targets of RNA, and the mechanistic roles of RNA binding proteins in RNA biology, gene expression regulation, and disease. RIP-seq allows researchers to gain insights into the RNA targets and regulatory roles of RBPs in various cellular processes, such as RNA splicing, stability, transport, and translation. By comparing RIP-seq data from different experimental conditions or cell types, researchers can investigate changes in RBP binding patterns and uncover potential regulatory mechanisms underlying gene expression. It has contributed to our understanding of RNA-protein interactions and has been particularly valuable in uncovering the roles of RBPs in various biological processes and diseases.
How does RIP/RIP-seq work?
RIP is a method that involves the immunoprecipitation of RBPs along with their associated RNA molecules. The principle behind this technique is to crosslink RBPs to RNA using a chemical crosslinker, typically formaldehyde, which "freezes" the interactions between RNA and RBPs at a particular moment in time. The cell or tissue lysate is then subjected to immunoprecipitation using antibodies specific to the RBP of interest. The immunoprecipitated RNA-protein complexes are then purified and the crosslinks are reversed to release the RNA molecules. Finally, the enriched RNA is analyzed to identify the bound transcripts.
RIP-seq combines the RIP technique with high-throughput sequencing technologies, allowing for the genome-wide identification and characterization of RNA molecules bound to RBPs. After the immunoprecipitation step, the purified RNA is converted into a library of cDNA fragments, which are then sequenced using next-generation sequencing platforms. The resulting sequence data provides information about the RNA sequences that were bound by the RBPs, allowing researchers to identify the target transcripts.
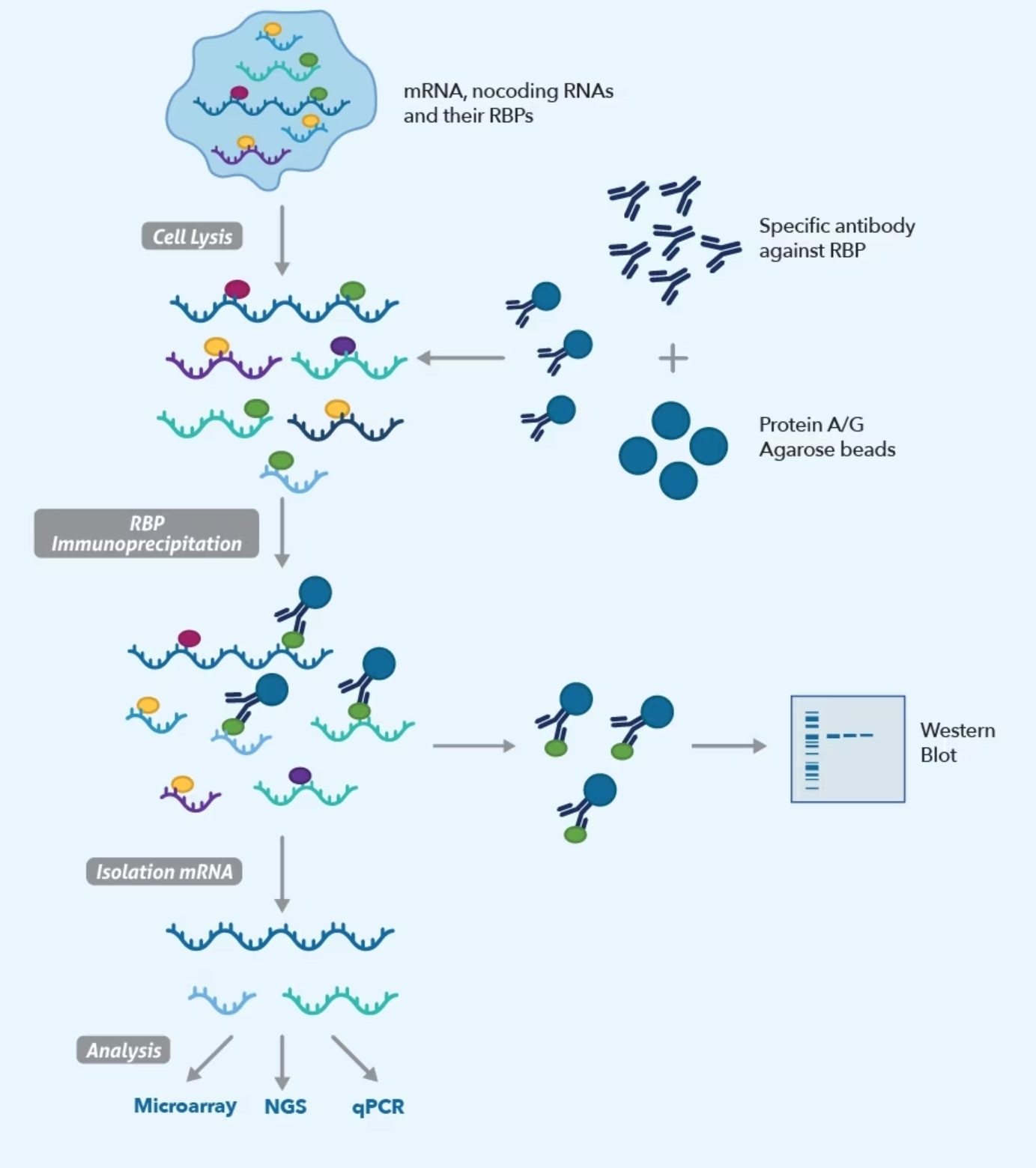
Fig 1. RIP/RIP-seq workflow
Here's an overview of the RIP/RIP-seq workflow:
-
Cross-linking: The first step in RIP-seq involves cross-linking the RBPs to the RNA molecules within the cell. This is typically done by treating the cells with a chemical cross-linker such as formaldehyde. The cross-linking helps to preserve the RBP-RNA interactions and prevent their dissociation during subsequent processing steps.
-
Cell lysis and immunoprecipitation: The cross-linked cells are then lysed to release the cellular contents. Antibodies specific to the target RBP are added to the lysate, and these antibodies selectively bind to the RBPs of interest. The RBP-RNA complexes are then immunoprecipitated using techniques like protein A/G beads, which bind to the antibody-RBP complexes.
-
RNA fragmentation: The immunoprecipitated RNA-protein complexes are treated with RNase to digest the unbound RNA molecules, leaving behind the RBP-bound RNA. The RBP is then removed from the complexes, leaving only the RNA molecules associated with the protein of interest.
-
RNA purification: The RNA is purified from the remaining protein fragments, contaminants, and cross-links. This step involves various enzymatic treatments and column purification to obtain high-quality RNA.
-
Library preparation and sequencing: The purified RNA is converted into a sequencing library by adding adapters and amplifying the RNA fragments. The library is then subjected to high-throughput sequencing using platforms like next-generation sequencing (NGS) technologies.
-
Data analysis: The resulting sequencing reads are aligned to a reference genome or transcriptome to identify the regions of the RNA molecules that were bound by the RBPs. The data analysis involves identifying enriched regions (peaks) that represent the binding sites of the RBPs, as well as downstream analysis to understand the functional implications of RBP-RNA interactions.
Proteintech offers antibodies validated in RIP to study RNA methylation
m6A Monoclonal Antibody (68055-1-lg)
 |
 |
|
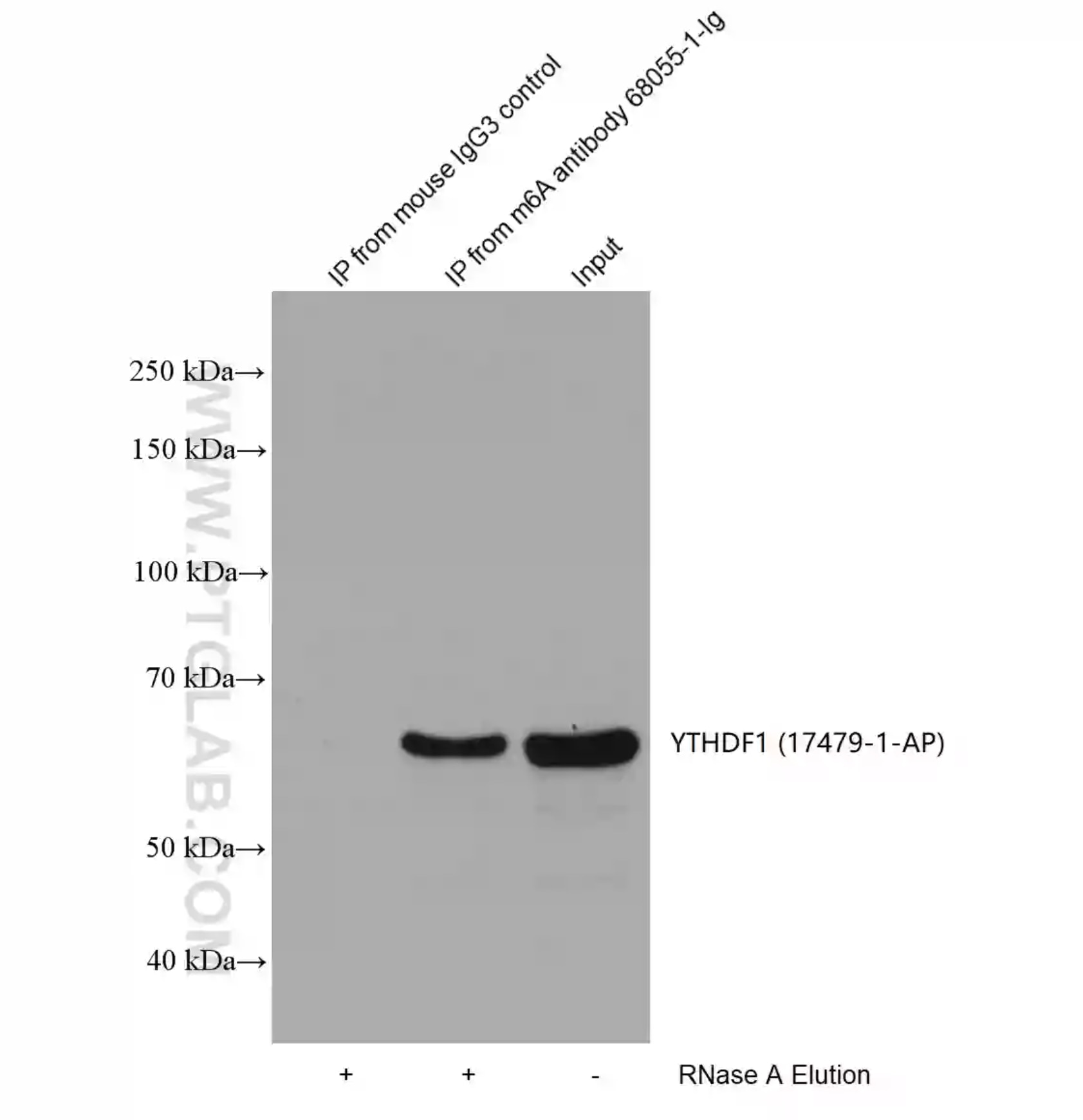
HEK-293 lysates were immunoprecipitated with Protein A-m6A antibody (68055-1-Ig) and Protein A-mouse IgG3 control antibody followed by western blot with YTHDF1(m6A reader) antibody 17479-1-AP. |
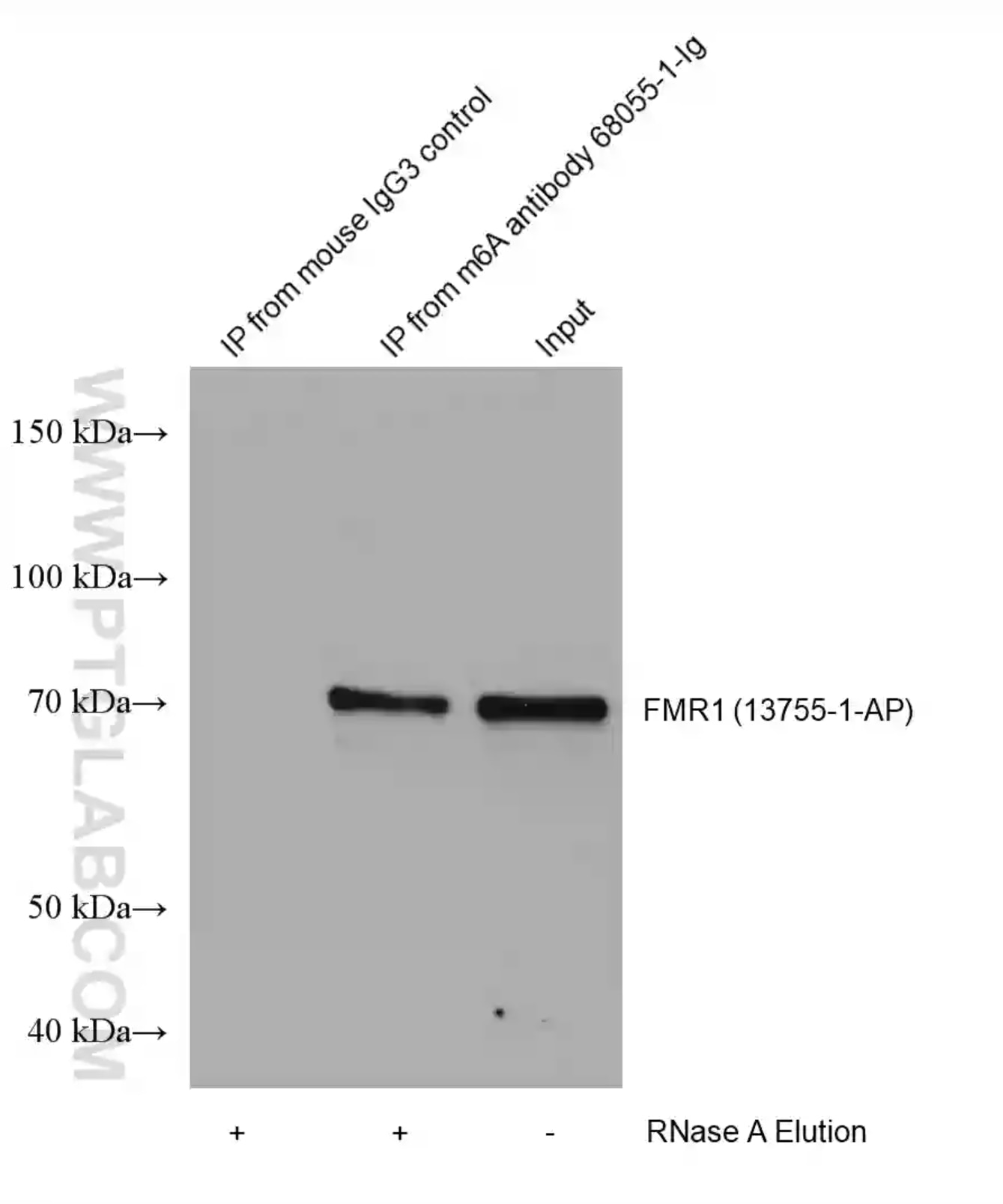
HEK-293 lysates were immunoprecipitated with Protein A-m6A antibody (68055-1-Ig) and Protein A-mouse IgG3 control antibody followed by western blot with FMR1 (m6A reader) antibody 13755-1-AP. |


